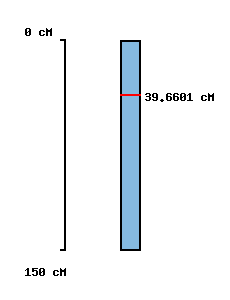
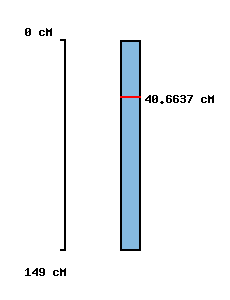
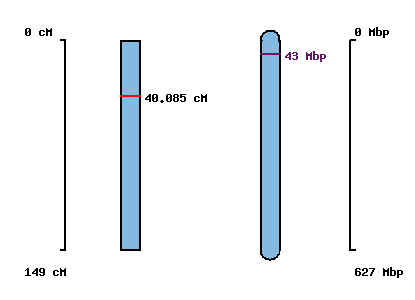
| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
 |
 |
 |
Predicted Genes from contig_9789
CDS Structure of MLOC_81723
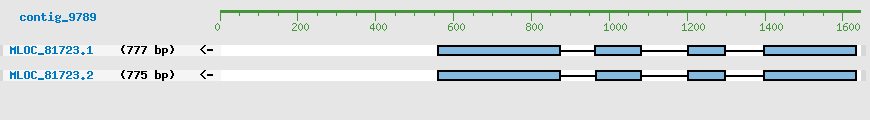
Contig Sequence (1644 bp)
>contig_9789 1644 assembly3_WGSMorex_rbca.fasta
GATGTTCCACTACATCAACCGTGTGTTTTTTCTTAACGCTTCTCGCTTAGCGNNCTACAA
GGGTATGTAGATCCGATCTTCCTCTCGTAGATGATCATCAGCATGATAGGTCTTCGCGTG
CGTAGGATTTTTTTGTTTTCCATACAACGTTCCCAACACATGATCCCACTGCCATAGAGA
TGGTCCCACTACAGTTTTTTTCTAGCTTTGTCCCTGCATTAGATCTGTCGCTAAATAACA
ATTCCCATGATTCAATGTAGTGCTCACGAGCGACTGTACTAGCTGATAACCCATTACACG
GACAAAGCAGATCCCCTAAACACTTCAAAATGCAACAAAGAAGAGGGATCATTGAATGGT
AAGTCTATCGAAGAAGGACCAACTGATGATGATATCTGAGGATGAACACCCAAAGCATCA
TGTATGACACTGATAGCATTCTTTTCGTTTTATTCACTATGCATATTATCAGTGGCATAA
GTCAAACAATATAAAGTTTGACTAAGTTTATAAAAGACAATACAAACGTTTACAAAGATG
AATAATGTGAAACTGTATTTCAATGATGAATGATGTGCTAGTTGAGCATAGTGGATGTTG
ATTTTCTTCAAGTATAAATTTTATCAAAGTTTACAAAGCTCAACTGCTAATATGCAGAGT
AAATAAAAAAGAGAGTACACACGTTGGGAGTAGAAATCATTAAGTAGGGATACATTGTGT
AGGCACACAAACAGCACAACTCCAACAGTGGAACTGGGCACGATCGATCCAACAGCGGAA
ATGGCGATCAGTTCTTGGACTTGGGCTGCTTGACTATCATCACGGAGCAGTCCGCGTGGT
GGGCGCAGTAGTCGCTCACGCTCCCGAGCAATGCCCTGCGAGAAAGAGGAGTTCAGACAA
GAGACAGTCACCATTAGTTTCAGACTCGCGGTGAAAGCGTACATTGGAGGAGAAAACCAC
GGACCTCCTGATGGCGCCGTAGCCCTGGCTCCCGACGACGAGCAGATCGGCGTGGTGCTT
CTCGACGGCGTCGCACAGGACGTGCTTCGGCTCCCCGTCGACCACATCCACCACGCCCTG
CACCTGCGGTTTCATATAAGCAGCAATGTGTCATCGTCTCTGTCACTGATGTCTTCTCCA
CATCTGTCCATGGAGGAATGCGACGAGCGGAGAGTTTGGTGGTGGATGTTATGTTATTAC
CGAGTTGGCGGCGCAGAGGCTGCGCGCCTTGTCGACGACGCCCTCGGCCTTCTTGCGCAG
GTCGGCCTCCACCAACCCCACCACGTCGCCGGCCACGCCTGAAACAAATTCCCAGAATTC
AGTCAGAGCTTTGCTCCAGTCCAGTCCAGAGGACGAACACGGAACGATGAAGCGGTAAAG
AGGAGGCTAACGCACCGGGGCCGCCCATGTTGACGGCGGTGGAAGTGGGCGGCTTGGCGT
GGATGACAACGAGCTCCACCGCTCCGGCGCCGGCCATGCCGGCCGTCGCCGCCACGTGCC
GCACGGCCCACTCGAGCGCGCGGTAGCTTGCTTCGCTGTCGTCCACGGCGACCAGCACCA
CCGTCTTGCTCCTCCCCTCCACCACCGGACTCGCCTCCACCGCAGCAGCCACCGTCTCGC
TCCTCCCCTCCACCACCGGACTCG




