| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
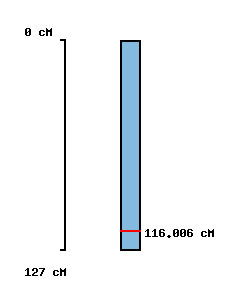 |
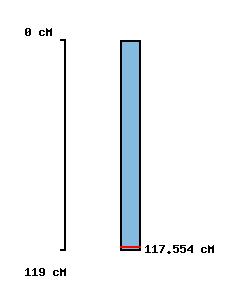 |
No map location |
Predicted Genes from contig_9118
| Gene ID | # CDS Predicted | Max CDS Length | Probes in Cluster | FPKM by Treatment | FPKM by Replicate | Top Rice Hit |
|---|
| MLOC_80539 |
1 |
299 bp |
None |
Available |
Available |
None |
CDS Structure of MLOC_80539
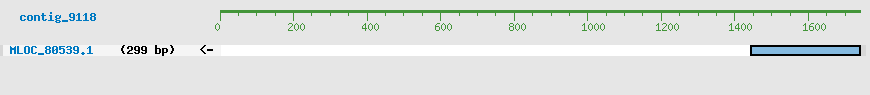
Contig Sequence (1740 bp)
>contig_9118 1740 assembly3_WGSMorex_rbca.fasta
GGGGGGGGGGGGGGGACTGGGTGGCATTTCTTATCCGGGTTGTTCGCCCGGACGTTTAGA
GTGGAAGTGGGGTGTCTTGTCGGGTGATATCGTGTGTCCGGGCTGTCCACCCGAATGTTT
AAGGAAGAAGTGGGGGTGACATCTTTTGTCCGGGCTATCTGCCCGAACGTTTAGAGAAAA
GGGGGGTGACATCTTTTGTCCGGGTTATCCGTCCGGATATTAAGGGAAGAAATGAGGGGG
GGGGGGTTGACATGTTTTGTCCGGGCTATGCGCCCGAACGTTTAGAGAAGATGTGTGGAA
GGGAGGGAGCGATCTAGATGCACTAGAAAATACATAGCACCTCAAACAAATAAAAGATTA
CATCGAAGACCTTGCACCGCCAAACAACCACTACCCCGTGAGAACTAGCCACCAATGCAT
CGTTGTCGCCGCTGCCCAAATAGAGCGAGCTTGACCTTGTAAATGACAAAAAGAGAAGTC
TTTGTGCGCACGTGCACTAGGGCCTAGCGCTCATCCATGAATCAATCTAAAGCGTCTGGC
ATGGATTTCATTGTTAGACAAGTACAACGAAAAACCCTAACATCACCGCCCCAATGGACG
ACATAAATCTACACCGAAGCTCTGCCTACTCCATCCCAATGGACAAAACTCAAGGAGGAT
CACAGACACAAGACTAACTTGACAATGAAGCTCCGCCATCCCCGAACTCTGCCCGCCCCT
ACAAGAAAACCCCCGACATAAACTACCGGTGAGAGCCGAGGCATCGGGAATCCCCTCTCT
TCCACCGGTTGCCTGATCGACATGCATAAGGGAGGTGGATCTATGGGCTCGCCAGTGAAG
CACATAAAGTTATATGTGCCGGAAAGCGTACTATTTGAAAGTTATGTACGCAAAAAAGCA
TACTATTTGAAAGCCCATGATATAGCAGAAGCATTTCATTTTCTTCTTGCATGTGTTGGC
GGTTTTGACCGATTACTTGACTTCCTTTTCCTCAAATCTTTCTCGGATGGCTTGGGATTA
CTTGGGCTCTTGGCCAGGCTAAAACCCTCGCCCGGCCTTTCGACGCCGGCAGCCCCGCGG
TTTGCCGTTCCCTTCTTGGAAGGTCCGCCATGGCCGTCATCCGCGTCCCGCTTCGAGCGT
CAAAGGAAACCTTAGGCCCCGATGATCTGGTCGGATGGCGACGACGTCGTTCTCCTTGAC
GAAGGTGTCGTCTCGTGTGATTGCGATGTCCTCGTTGGTGGACTCGGAGATGTTGATATT
GCTTGGGCCGCTTACGGGACAACGAGTGGGCTCGACATCGCACGTCTCTACGCTCTGCGC
ATCGTGTTTACGCGATTCAAATATAATAATAATAGGTCTGATCTTTTTAAATAATATAAA
TGAATAAAAAACTCAACGGAGGTGGGATATATATGCAGCCAAAATCTGTTACCACGAGTG
GCACGTCTGATCCGGTCTGAACGTGCGCGTGCTGCTGTGATTAAAGGTTGCGTGACGTTA
CGTGCGAACAGCCGCTTTCGCTTCGGCCGCTTCCCTTTTCGATAAGAACCAACCAACCAA
CGATCTCCCTTCCATCGCTGCCCAGCAAGAACACGCCTCCCTCCTCCGACCCCGAGCTCC
ACAGAGTCAGCGCAAGGGGATCTCCCTCTCCCTGCTTCCTCCATGGAGAAGGCGATCGAC
CGGCAGCGCGTGCTCCTCGCGCACCTCCTCCCCTCCCCCGCCTCCGCCGGCAGCCAGCTT



