| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
| No map location |  |
No map location |
Predicted Genes from contig_442860
CDS Structure of MLOC_60336
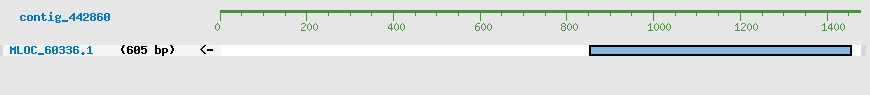
Contig Sequence (1476 bp)
>contig_442860 1476 assembly3_WGSMorex_rbca.fasta
TTATATACCGAAAACAATTTGGCAGGTTTTCTCTTGGCTGTATGCTTTTTTGCCAGGCTT
GTGTGAGATATATACTGTTGGCTTAGCTACAGTTTGCTGGTCTATTTGGTTGGCTCGGAA
TCGGGCTACATTTGAAAAAGAAATGGACTAAAACTCCTTTTGAAATTGCTTTCACAACAT
GTGCGTTTATTGAATACTGGGCAGGTATGCAAAACCCGGCGATGACAGGAAATGTCAAGA
AGGGAGCTCAGTTGCTGAAAGAGAGCGCAGCTCAAATGATGCGGCTGTGTGGACCACCAA
GGCCAGAAGCGAGCGAGCAGGCTGNNGAAGAAGAAGTCTGGGATGAATGGTAAATGCTGG
AAGGCTCTAGGTCCAGTTGATGATAAACTTTGTTGGTTACAGTGCTGATGGCTTGGCTTT
CATGGTGATGTTTAGGTTCTATATCCAGTTGGTGAAAAGTTTCGCCTTATGTTTGTGCTC
TTGTGTGAGCCTGGTTCGGTGATGGTTGTTTGGGGGCTCCTGGTAGTGGTGCTGATACAT
GGCAGTGTAATACTTTTGGTCTTTGTTGCGTAATTTTGGATGATATAGGTTTTCGGCCCG
CAGTTTGTGTCCCTGATGACAGGCATGAACGGTGGGTTTCCTGATATTGTCCCGTACTCG
CTTTTTTCCCTTTCCCTCTGTTTCTGGTTGGTTCTGGTATCAATCAAAAATTGTATTTTT
TCGTTATGAAAGTAATGGAAAGGGGAGAAAGCCCGGTTCGAAAAAAATGGAGACGGAGTT
GTATGGCTTTGGTCAGGTATCTCGAAATGTAAACACCAATTTAGTGCAACGTATATATTT
CTTGATTAATCGACCTGTGTGGGCTTGCATTCCTCACTCGCTACGACAATGCAGTAACAT
ATAAAAACAACATCTTTTGTTCATTCAATATAAGCCCTTTTCTAGTCTATGATCATCTTT
TGTTTTGTATAAGCCTGCTATATATAACCAAGTAAATAATAATCAGAATCAAAGTAAAAG
AGAAACAAAATACAAATTTAAAAGAATATCCGTGTCCTAAATATAGGGATTTGTGCAGCT
CCTGGAACTTTTTGTGTCTGTATGTCCATCCAATCAATCCATTATGCTGCTCGCAGTCTT
CTTGATGAGGTCCCATGCACTCCTCACATGCCTCTCCTCCTGCAGCGACGAGCCAACCGC
AAACCGGAGCACGAACCTGTCACCGACCACCGTGTGCGCAAGATATGCCTTGCCCGTTTT
GTTCAGATTCTCCATTAGCACACGGTTGGCCTCGTCGGCGTCCTCTTCCGTCATGGCTCC
ATTTGGCTTGATCCTAAAGCACACAAGAGCAAAGTTCCTCGGTACGACCACCTCAAACCT
GTTGTCGGCACGGACAAAATCTTCAAACATCTTAGCCATGGAAACATCACTACGGATGTG
TTCTTGAAGGTTTGCGGTACCATAGGTGCGCATGAC


