| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
| No map location |  |
 |
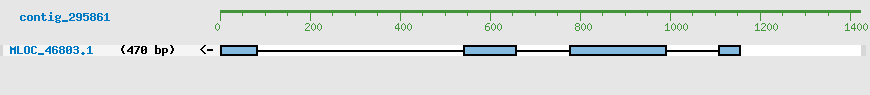
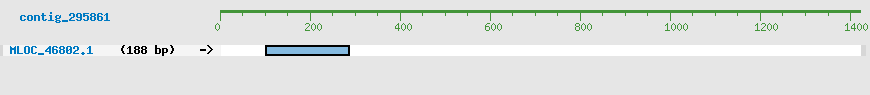
Predicted Genes from contig_295861
CDS Structure of MLOC_46803
CDS Structure of MLOC_46802
Contig Sequence (1421 bp)
>contig_295861 1421 assembly3_WGSMorex_rbca.fasta
GCAGGAATGTAGTGCCCAGCATATGACTCCCCGGTTATATAGAAATCGTTTTCAACATAC
TCCGGGTGCTCCTTGAAGAAGGCCTATCAAAATTGCATGCAGAAAGACACAACGAACTTA
TGAGAAAAAACACGGAAGGCCATTTTTGCATCTTTATTGCAACTCAAAAATAAGGGTAAT
CACTCTGTTTCACCTCAACTCTTGCATAACTAACCTTGTCGCCACAATAGGAAGCACATT
AACAATACATATGATGCATGTGTATTTCCTACTACATATGCTTACCGACTGGGAATGCAT
ACCATAATATTTCAAGTCCATAATACTTCCTCCGTTCCTAAATACAAGTCTTTTTAGAGA
TTTCATTACGGGCTACATATGGATATATATAGATATACTTTAGAGTGTAGATTCATTCAT
TTTGTTTCCTATGTAGTCCGTAGTAGAATCTCTAAAAAGAATTTTATTTAAGAACAGAGG
GAGTAGTGAACAAGCAAGGGCTTATCTTCATTAATCACAACAGGAGTAACTATAATTACC
TGCGGGAAGTCATATAGGTCGTTGCTCACACCCACTTCATTATGGTGGGTATCGTGCGAA
TCGGAGCTGTAGTTGAACCCAGTCCCAGTTGGCTGATCAACATAGATAAGATTCGACTCC
TGCATCAGCCAATATCCAGTTCCAAACAGAGAGAAGATTAATCATACAGGGTTCTAAACC
AACGATCGAGGAGCAGAATGCAGTATCCAAACAATTGATCTGTAAGAAAAATACCACACG
GATTGGTTGATTAGCACACAAACTGCCTGGATATATCTACCTGATCCCAACCAAACTCAT
TCCAGAGCAACGACATGTTGTCCGCAATGTGGAAGGGGCCATTCTCGTAGAAGAGGGCTA
GCTCGTTGTTGCATCCGGGCCCTCCCGTCAGCCAGATCACCACGGGGTCCTCCTTCTTGT
GTCGCCTGGATTTGAAGAAGAAGTAGAAGAGCCTACACGCAACATGTACAGTGACAGATG
TCAAAGATACAGGAACAAGATCCTCACGGGCAAGGCAGTGCGAAGGAAGATGCCGGCCGA
AGACTGGACGAGTGAGCTCGCTCACCTGGCGTCGTGAGTGGTGGGGAGGCGGTAGTACCC
GGTGTGGTTCCCGAGGTTGCTCACCGACGTGTTGCCGCCGCCGTCGTCTCCGGCCACCAG
GGAAGCGAGGCGGATCGGCCTCTCGGTGATGGTTCCGGCCGGGAGAGCGTCACCTCTGCC
GACGCCAGCGTACCGGTCGACGGGGTCCTTGAACTTGGAGAGGAAGCACTCGTGGTCGTC
GAAGACCGAGGCAATGCACTGCTGCGCGCCCAGCTTGTTCGCGTTCACCTCCTTGAGCTC
CCCCTCCGGCATGGCCAGCACCGCCGTCCGCGCGCGGTCGT




