| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
| No map location | No map location | 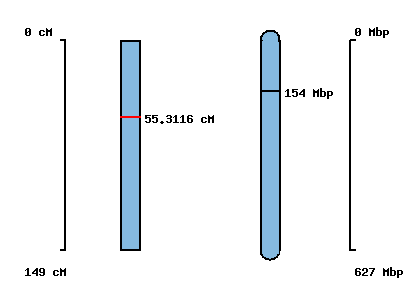 |
Predicted Genes from contig_280637
CDS Structure of MLOC_45739
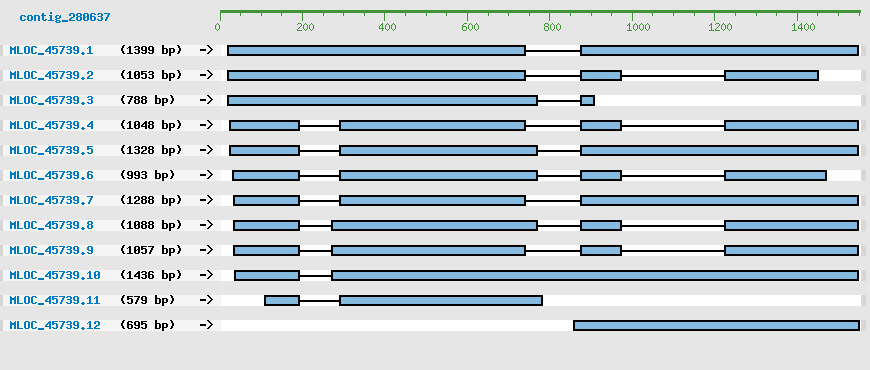
Contig Sequence (1552 bp)
>contig_280637 1552 assembly3_WGSMorex_rbca.fasta
CAATCCCCGCCGGCGGGCCGCCCCCCCCACCCGCCGACGGATCTCCGGATCTGTGCGGTC
TCGCCCCCCACCCGCCTGGATCCGCACCGAGGCGCGGGTGCTGCCTCCATAGGAAGGAAT
TCAGTTCTCCCCCTCACAGGCGGCGTCCTCGACGGTCAGCGGCGGCGACCCGGCACGGGA
GTCCTTTCGCCGGTAAGGAACCCTAGATTTATGTTCGTTTTCCTCTTCTCCCGTGCCTTC
CTTTGCTATGAAATGCCGAATCCGACCAGATCTTGAGTCTTGCCACAGGCCGCTCTCGTC
GAAGGGGGCCTGGTCGAGGAATGCACCGTACACCATCGTACACCGTCATACAAGTGCAGA
ACGAGCTGAAGCAGAACAGGGCTTCCGACGGCCATGTGATTGCCGCTCCGGAGAAGGAGG
CCCTTGCTGCCGGTTGCCGCAAGGAGCTCGCGGGCTTGTGGGCGTACTTCAAGGAGGTTT
CTGGCCACAAGATGCAGATCGATGGTGGTGGAAACCTGTCAAGCAATGCCATGATTGGGT
GCTTGCTGGAGGAGAGCAATCTTGGGCTATCCAAGTTGGTGGATGAGACATTTCAAAAGC
TCAAAGGAACAGAGGGTGTCTCTGTTGCTTCGGTCCGCAGCTCAGTCCTGCTCATTGGAC
AGAGGATGATGTACGGACAATCTAGCCCCGATGCTGATGTGTTGGAAGATGAATCGGATC
TTTCTCTTTGGTGCTCGGAGGCAAATCCTATCCCATTTTCTTAATTCTGAAGTTGGTGCC
ATAGCATTTCATTTTAATGTTGTTTTAATCCAGAAAGACCGGTTCTGTTCAAATTAGTAT
GTCATGATGTCTTGTGCTGGGTTGTAATTGCAGGTAAGAGATTTGAAGGTACTGCCTATG
AGAACGCGTGGGTTTTTAAGTGTGCGGAGAACTGCTAGAAAGAAAATCCATGAGAGGATC
ACTGCTCTCCATTGTAAGACATATCTTCTTGCAGCATGTTTGAATCCCATCTATCAATGT
TTGACAGTTAATATGCGAATTGCTGGTTTTTTGGTTTGTATATCTCAACTTGTGCCCATT
CTTTGAATATGCAACATGCTTTGTTGTTAATTTTTTCCTGAAATGTCATTGCAAGTTAGC
TTTCGTTGACATCTAATTAGAACTGTGAATTTGCCTACTCCATGGCTTTGTATTCTGTTC
ACTAAGTGCATCTCTATTGCAGCAACTTTGTCAGCTCTGGAAACTACAGGAGCTGAAGGT
CAAGTAAACGAGCTTAGAAAAGTTCTCTAGGAAGAGAATTTTGTAATATGTACTTGTTAA
CTGATAGTCATAGTGACTATCGTGCTACTTAGAGTGGGGGACACCGTCTTCTGGGATCTT
TAGCCACAACTCAAAAATTAAAAAGGAAGTGTTTTATCTATTCTACTCGAGTGTATAAAT
ATTGCATAGTGTCACTTAGAGACAGGGTAGTCATTCAACATTTAGTTCACCAGATTGGGA
CAACTTTCCTGCTTTCTTTACTTTGTTAGTGTGCCGTTTTTGTCAGTATTAT


