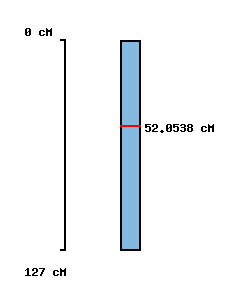| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
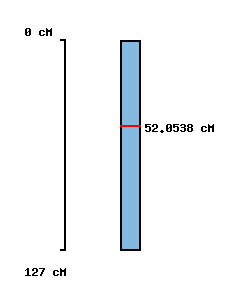 |
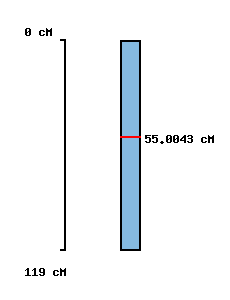 |
No map location |
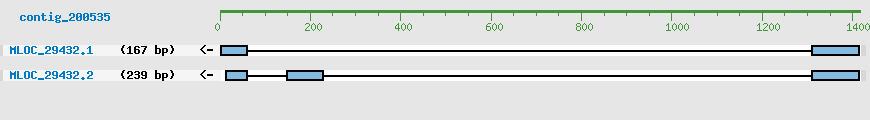
Predicted Genes from contig_200535
CDS Structure of MLOC_29431
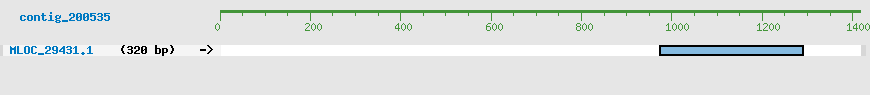
CDS Structure of MLOC_29432
Contig Sequence (1416 bp)
>contig_200535 1416 assembly3_WGSMorex_rbca.fasta
TCGCCGAGCAGCCTGTGGGCCGACTCCCTCACCAGGCTCGTCTCCTCCAGCAAGTTGTAA
TCTGCACGCACACGGAGAAAAAATAGGGGACGATGAGCATGGACGGAGCGGGGGACTGTA
CATGGGGTCGGGGATGAATCTGCTCACCGGAGAGGAGCTGGGTGTCGTCGAACTGGGCGA
GCAGGACGGGGGCACAATTAAATGATTCATCAAAGAATTTACTCATATCCCTAACATCCA
TTTGCATGTCTACAGAAATCAAGAGACACTAGATTTCACTTCCCGAACAAAGAGGGAATT
TAAAATATTCACTAGATTTCACTTCCCGAACAAAAAGGGAATTTAAAATATTTAAGGATA
CATTGTAGCCATCAGCGACCATATCCTCTGACATAACTGGAAACATCAGAGCATGGGAAG
AAAATTTAAATTAGCACAGTGAGTGTAATCATCGAGCTAGGTTTAGAAGCGCATCCTGAA
CTCTATCCAGAAGACCAGAACAACCCATTGAGGGTGCAAAACACAAGTAGTGAAAATCAG
ATGTGCTGCCGTGTGTTTAACGTGTCGATGCAGTATTGACGCCTGCACCATAATACTAAA
TCCCAGAGCAACTTTGGGTTATTGGGGAACAAGTCAGATCAAGGTTGGTTTTAGTTTGGA
TTATTATCAAATAGAGATGTGCTGGTTTCAGATGTGTGCTTCCATATTACTGTCCCTACC
CCATTTGGCCATGCCAGTTTGCAATGACTGCTTTTGTGTGGCATCAACATCACAACCCTA
TCAATTCGCACATTGCCAAACTGTAGCAGCATAGCACATTACCCTATCCAGAGCATAATT
ATCAGATTACTAGAAGTGATATATGTCAGCATGTGGACTGATAGAATTGCATTATCTGAT
TGCCATGTCATCATATACAGAACAGATACAGGAAGCACTGGAGCTAAATCACCACCTAGA
TGAGGAGCTATCCATAAACTATTGCCGTGACAGCAATTTCGACCCACAACATACGGTGAC
TGTGCTCTGTGCAAGCAGCTAATCCAAAAGATCACCCGCTCGCTGGTCCATGCCAATCAC
CCCCATCCCTTCACAAATCACAATCAAGCACCTATTTGAACCGAACCTGAAAAGGCGGAA
AGGGAATTCCTACTTCCGGAAAAGAGAGACGAATGCACAAGCAGTCAAGCACCACCACCA
CGAGCACAAATCAGCTACTCCAGTTCGGGATCTAGGCGAGCGGCAGTCTCGTCTATGCGG
AGCAATCGTGAGGAGGAGAGCAGAAAAGGGGAAGGGGTAGGACGTACGGGCGGACAAAGG
GGCGCAGCGCGGCGTAGCGCTGGTACCAGTCGTAGGGCGCGCCGCCCTCCTCCGCGTAGC
GTGCGTCCCGGTACCCCGCCTCCCCGTAGCTGAACA