| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
| No map location | 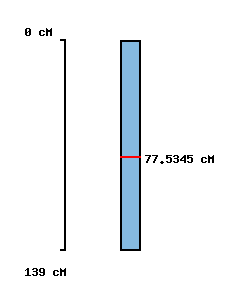 |
No map location |
Predicted Genes from contig_1966601
CDS Structure of MLOC_29015
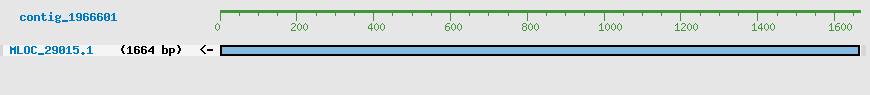
Contig Sequence (1666 bp)
>contig_1966601 1666 assembly3_WGSMorex_rbca.fasta
ACGGAGCCGTTGAGCATGACTGCCGATGTCGCGGAGCTGAGAAGGAGAGCTATCCAATCG
CGCCAACGCGAGGGGAACCCTAGCCGCTGGAGGAGCTCTATCAGGTACTCCCAAGAGACG
TTGTCAAACGTTCTGGCGATGTCGAGCTTGAACATCAGCGCGGGCCTCTTCTTTCGGTGC
AGCGAGCGTACTGCGTTTTGTACGTACAGGAAACTATCTTGTGTTGACTTGGAGCTCACG
AAGGCGGATTGCGCCGGGGAGATGATGCTGCTGATGACCCCCGATAGTCTGGTAGCTAGA
ACCTTGGCGATTATCTTAGCTACTGAGTGGATTAGGCTGATCGGTCGGAAGACCTGCACC
TTCATCGCGCCTTCTTTTTTTGGGAGGAGTACAATCATGGCCGTGTTCAGGCTGCTAAAA
TCGCCCCTGGCCAGGTGGTAAAAACTATGAAAGACTTCCATGATGTCCTGTTTGATGGTA
CCCCAGCAGGCTTTGTAGAAAGCCCCTGTGAAGCCATCGGGGCCTGGAGCCTTCTCCGAC
GGGGATGCCTTGATCGCCTCCCAAACCTCCTCCTCCGTGAAGGGGTTGTCCATGNNNNNN
NNNNNNNNNNNNNGCATGTCGTGCCTGTCCCAATTTATGCCCGTGTGGCGATCTACCGTG
GCCCCAAGATGGTTGCAGTAGTGTTGGTATATTGCATCTTCTTTGTGCTCGTGCGTTGTA
GCTATCTCCGGTCCCACTTGAAGGCAGTGTATGAAGTTTTTTCTATTTCTTGCTCTTACT
TTGGCGTGGAAAAATTTAGTGCTTGCGTCACCTAGCCGTAGCCAGGTGAGTCGGGATGCT
TGCCGTCGGCGGGCACGTTCGAGAGCCACTAGGCCTAGTAACCGGAGCTTGAGAAGCCTT
CTTAGGTTGAACTCAGCGTCCGTGAGGATGCGTTGTTCTTGTGCTACATCTACCCTTAGA
ACTATCTCTGTTGCCATATGGAACTGGAGCTTGGCATCACTGAAGAAACCTTTGCTCCAG
ATTTTTAGGTCCCTGGCGGTGCGTACCATCTTAGTGTTCAGCCTTTGGAAGGCGTTGGTG
TCTTGTACTGGTCGATTCCATACTCTAGCGACCGTGTCGTGGAATCTGGGGAACGCCGGC
CAGAAGTTTTCGAATCTAAAGATCGCCGCCCTAGGTACCGTGCTGGTGTCTGCTAGGATC
AGGGGACAGTGATCAGAGGTGGCCGAGGCTGCCGCCTGAAGAGAGGCTGATGGGAACAGC
CCTTCCCATGCTTGGTTGCAGAAGAACTTGTCTATGCTCATCTGTGTGGGATTTCGACGC
TCGTTTGTCCAGGTGTATCTACGGTTGTTACATCTGATCTCTCTCAACCCTGTCGTATCT
AAGGCTGATCTGAATCGGCCCATGAGTCTGCGATTTAGATTTAGGTTGTTTTTGTCCCGT
GCCTCGTAAATGAGGTTAAAATCTCCGTTGATAAGCCATGGCTCTGCTGCAGGCGGAGCA
GATCGAGAGAGCTCCAAGAGAAATCTATCTTTTCTAGCGTCATCCGCCGGCCCGTAGACC
GTTGATAGCCAGAAGCTATCGCGTTGGCCAAGCAGGCAAACCTTAGCCGTGATTGCGAAC
TCTCCTATGGCGTGGGAGGTTACGGTAGCAAGGTCTTTGTTCCATA


