| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
| No map location | No map location | No map location |
Predicted Genes from contig_192579
| Gene ID | # CDS Predicted | Max CDS Length | Probes in Cluster | FPKM by Treatment | FPKM by Replicate | Top Rice Hit |
|---|
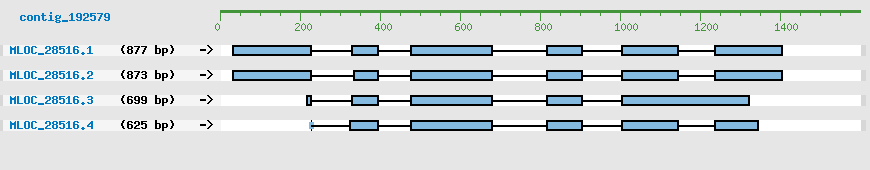
| MLOC_28516 |
4 |
877 bp |
AK250051.1 |
Not available |
Available |
None |
CDS Structure of MLOC_28516
Contig Sequence (1598 bp)
>contig_192579 1598 assembly3_WGSMorex_rbca.fasta
CCACCCTCTCCGCCTCTCCACCACCGACGCGCCATGTTCGTGTTCAGGGGGAAGTCCGTG
TTCAACCTGAGGAAAAAGTTAGCCCGCTGGCTGGAGGCCCTCATGGACGTCTCCAACCTC
GCCATGTGCGTCGAAGCCGGTACTTTCGAGGTTCTTACCCAGATCGTCGTCGACCTGCCC
CGCAGCCGCCAGGAACTCCACATTGTCGTCTTCGCAGGTGGGACGCCAGGTGAGCCGTCC
ATGTCCTAAACTTCCTACTGATCTGATCTGGTGTTGGGGAACAGTCTGAAAACAGTTTAC
TGAAACTCGGACGAATCTCGCTGAAAAGATAGGCCAGATGACCCAGATTGAGAGGGAGAA
TGCCACAGCAGAGGCCATGTCCAAGTACTTCATAGGTGTGTCTCGCTTCCCTTGCGCTAA
ATTAACAACTGCAGTTTGTGAAGGGTTGTGTGTAATAACATTTTGTATGATGCAGGTAGC
GTACTGAGTGGTGGAGGTAGTGTGCCTTCTCCTCAAGCATCCGCTACGGCTTGGCAGTCG
ATGGTGAATGAAATGCAAAAGGGCGCCCTTTCTACCCGCCTAGGCATTCCGATGATCTAC
GGTATTGATGTTGTGCACGGTCACAACAACGTGTACAAAGCTACTATCTTCCCACATAAT
GTTGGCCTGGGAGCTACCAGGTAGAAACAATACTCTTTTTTAACTTTTGATGTTCTGAAT
TATATCTGATCAACCACCTACAACAATTTTCAGAAAAGGCATGCTAACTGTGACACATAT
GTTGTTCTGAATTAATATGTTAATGTCTTCTAGGGACCCTATGTTGGTAAAGAGGATAGG
ATAAGAACCTGCTCTTGAAGTTAGAGCAACAGGAATTCCTTACGCTTTTGCTCCGTGTAT
TGCGGTAATATATTGTACCTCTTGCATGGTTGATACCTTACTATGGACAGAAAATGTGCC
TTCTTATTGTTCCTAAGGAACTCACCTGTTGATGTTGCTCAGGTGTGTAGAGACCCAAGA
TAGGGACGCAGCTACGAAAGCTACAGCGAAGACCCAAAGGTTGTCCAGTCAATGACCATG
CTCATCTCTGGTATGCAAGGTGACGTTCCGGCTGGTTCTGAGGGAAGGCCATTCGTTGGT
GCAAGGTAATATACATTCAACAGGGAACTTGTGTTCTGTGATTTAGTCTGAAGAGACTGA
GCTAACTTCATTGTCACTCTTGGTACACTGCAGTAAGAAGGTTGCTGCATGCGCAAAGCA
CTATGTTGGTGATGGTGGTACGTTTATGGGGAACAACGAGAACGATACAATCATTGACGC
CCATGGGCTGATGACTATCCATATGCCTGTTTATTACAATTCTATCATCAGAGGTGTCTC
CACTGTTATGACCTCGTACTCTAGCTGGAACAGAAATAAAATGCACACCAACCATTTCCT
TGTCACTGATTTTCTGAAGAACAAGCTCAAATTTCGGGTGAGTTGCTCATTAGAAAGCCA
GGAGAATTTCTCTCTCTTGTTTTCTTCAGACATATTTAAAAGATACTGATAAAATCAATT
TCCGGGATTTCCTGCAAACAGGGTTTCGTGATTTCAGA

