| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
 |
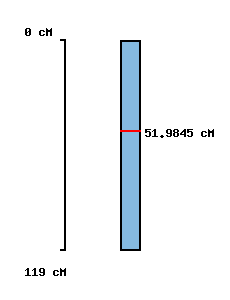 |
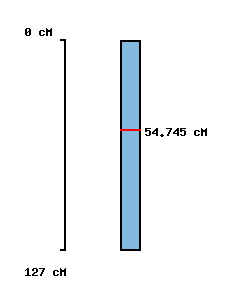
No map location |
Predicted Genes from contig_181189
| Gene ID | # CDS Predicted | Max CDS Length | Probes in Cluster | FPKM by Treatment | FPKM by Replicate | Top Rice Hit |
|---|
| MLOC_27178 |
1 |
1386 bp |
AK365584 |
Not available |
Available |
None |
CDS Structure of MLOC_27178
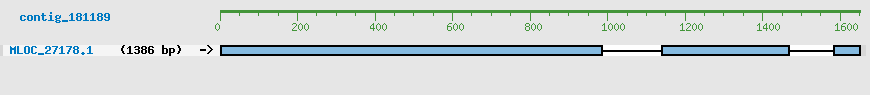
Contig Sequence (1651 bp)
>contig_181189 1651 assembly3_WGSMorex_rbca.fasta
CTACTATTAAAGGGGGATCTACCGTCATCATGATGGTTCGACCTCCTTCAATCTCCCCCT
CCCAACGCTAACGCTGTCTACCCTCCCACCCATGTTTCACCACCGATTTTTATTCAACCC
TTCAAAAACCAGTCGGTTTAATTAAAAAAAAATCTCTTTCTCCCCTCCTGCCATCCCACG
GAGAAAAGAAATCGCAAGGGGAAAAACACGTAAGCCTGCTAGGATTCCCACCTTCCCCGC
ATGTAAACCAACGACGATGAGGCACCCACGCCCCATCTCACACAACCCCTCCCTCCTCCC
CATCCCATCCGAGAAAACTCCTTGGCCGTCCAGCCCAAACTGCCATGCCTGCACAAGATC
CCGTAGTCCCTCGATCCAATCTTCGCCCTCGCCGACGCGGGCGGCCCCGCCACTCTCCCG
AGAAAAATCCATCCAAGACCAAGCCCGTGACCAACACCCTCACGAGAAGACCGAGCCCGC
GACCAGCTACGACTTCTTGATGAGCCCCCCCGGGACTTCAGCAACAACATCTTCAATCGG
AATGCCGCCAAGCTCGCGCACATCGTCTCCGGCAATGCAGCTGCGAAGCAACCCATGGAC
TTCCAAGTCAACCTGGCAGGCTCTGGGTAATTGCAAGACCATCCTGTGACCTAACCTAAC
ACTGTTGATTTGTGTATATGTGTTTCTTGGATCAGGCCTTGCTGATGAAAGCGATGACGG
ACTCCTTATTCACCACCGCCTTCGGCGTGGACCTCGCCACGCTGTCGGGGTCGGACGACG
GGAGGTGCTTCGCCGCGTCATTCGACGACGCCAGCAAGTTCATCCTACTCCGTTACGTCG
ATGCATTCTGGAAGGTGTCGAGGTTCCCCAACGTCGGCGCTGAGGCGGCGCTCAGGCACA
GGATCAAGGTCGTCGACGAGTTCATCTACAAGCACATCCGTGCCAAGGCAGAGGAGATGT
CGGCTCGCGCCAAGGCACACGACGCTGTATGTGATCCAACGTGTTCTGAATTTCTGACCT
CCATCAACTTGTTGTTACTTGTTGTGTTGCACGTACATACAGTATATAGTACTATACTGT
ATTATGCACGCTTGAAACTGAAGCTCAGGTTCTCCTTGACTACGTTCATAACTTGATAGG
GGTCAAAGGATGATCTGCTGTCCAGATTCATATTGGCGACCAACAACGACACCGAGAAGG
TGGATTACAAGTACCTGAGGGACATCATACTGAACATCGTCATGGCCGGCAAGGTCACGA
CCACCGAAGCGCTTGCTTGGTTCCTTTACATGATGTGCAAGAACCCGGAGGTCCAGGAGA
AGATAAGCAAGGAGGCCGCCGACGACGGCGAGGCCACGTCGTCCATCGACGACTTCTCCT
ACAGCCTCACTCACGAAGTGCTAAACAAGATGCACTACCTGCACGCCGCCCTGACGGAGA
CGCTCCGGCTGTACCCTTCGCTCCCGCTGGTGAGTCCATCCATCTACCATGAGTCGACGA
CACACTCTAAACTTGTTGGGATTCCATGTAAACATCATAACTGCAGATCTGAGTTCTTCT
GTATTTTTTGGGGTGTTTCAGGATAACAAGGAGTGCTTCTCAGACGACGTTCTGCCCAAC
GGCTTCAACGTCGGCAAGGGAGACATCGTGT



