| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
| No map location |  |
No map location |
Predicted Genes from contig_1635225
CDS Structure of MLOC_24040
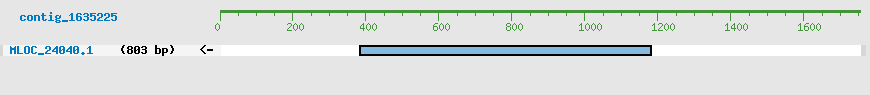
Contig Sequence (1754 bp)
>contig_1635225 1754 assembly3_WGSMorex_rbca.fasta
GGGGGGGAGGGGGAGTGGAGATGCTATAAATGTCTTCCACAGCCACCATGGGGCTGATCA
TGTCGTGGCGCAAAGCTGGTCACCGTCGCTCCACACCACCGCGAGCTGACATGCAACATC
CATCATATCCCTGGGGTCGCTGGCCCGGCATCCTCCATGTGAAAAGAGACCGATGTTGTT
GGAAATATGGCACAACATCAGTGATATCCAATCAGCAGCGCTTGCAAAAACATCAAAATC
CTGTTTGAAATATTGACGTATGTGGATTTGTTCAAGTAATAGTTAGAAGTGCATGGAAGT
TCTGTTTTGTGTTTCATAATCCTTCTGCTGAAGTGCTGAGGCAATCACTGTGTCTGGATA
TAGACGATTTCATTATGAAAATCAACGGAGAAACTTGTTATTTCAAGTAAAATCAGACTA
TACAAGAAGTATCTGCCAGCCTCTACTTTCCTTGAGTCTTAAATTTGTTCCCTTGGGTCC
TTGTTCGACTGAGCCTTCTGTGTTGCAAGCTACTGCGTAGTAAACAGAACAGAACATCCT
TTTTCTTTCGAAAAAGGGATCACCCGGGCCTCTGCGTCACTGGATGCATACAACCAACAG
CGCAATCCATCGCATAAGAAAATTTACATAACCTCTCTACTCCCTCGTACAATCCAAAAT
AACACTAGTATGAAACGGTTTCTCACCTAATTCAGAACCCTGCAGACGCATAGGCCGGGG
TCTTCTTTCTTTTCTAAAAAAAAAACCTAATTAAGAACCCAAGGAAGGTCTGAAGGAATG
GGAGGAAGTTAACGTGTACAGCAGGTACAGGTACTGCTCTAGTCTAGACTCTAGAGCCTT
GACTGAAAATCGATGTGGTTTGGAGAGGTTCAGTTGTAGGTGCCGAAGCCGGCGCGACCT
GCTGCTCGCTTCTCTGCGCCCGCGCCGTCATTCCTCGGCAGCACGAGGTTGCTGATGGTC
CTGATGATCTGCCGCTTCAGCTGCCCCCTCTTCAGCGGTGGCCTCTTCTCCTTTGTGCCG
CCGTGGTTGCTGATGCCGCTGTTGGTGTGTTGGTAGTACTCCATGGCTTTACCTGTGTTG
CCCTCCCTCCTCTTCTTCTTCTTCTTTGTTCGTGTTTGTGTATGTGTGAGAGAGAGTGTG
TTTTCGAAAAGGACATGAGAAGTTGAAAACTATTTATAGGCGAACTAAATCATCCGTGAT
GTGTTCATACTTCGTACCGGTGCCTTGCCGCACGTGTTATTGGTTCAGTTAGGGGGAATA
AATTGACCGCTTGTGGCCCACGATGTTTGTAGCGTGAAGGCACTAAACAAAAAGAAGTTC
CCCACACGAGTACTAAGCTCCCCAAAACGACTGGAATAATTGATTATTATGGCAGATTGC
CGGGTGCATCTTTGAAGCATCCAGAGACTTCTGACCGGTCCATCCATCCAGACCGATTTC
GAGCTCATGGAAAATAAATATCCAGTTCAGCGTGTCATTTGGACGTCTCCGAGTTTTAAA
AGCGAGAGAGGAGATTGGATTGGATTCATCCCCCCAACGTTTACAACTCGTCGAACCGAA
CTGAACCATCAAGCACTAGATAGATACTCCCTCCGTCTCTAAGGGTGTGTTTAGTTGCTG
TCGAATGTCACGGCCGGACCCGTTCCTAGTCCTCGTTCCTGTGTTTGGTTTATCAACTGC
AACAGAACCCACACATTCCACTTTAGGGAATATTCCACTCAGATCCGGAACGTGCTAGTA
CCACGAAATTGATG


