| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
 |
No map location | No map location |
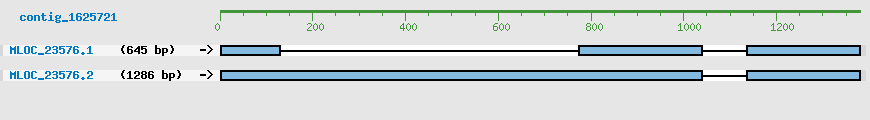
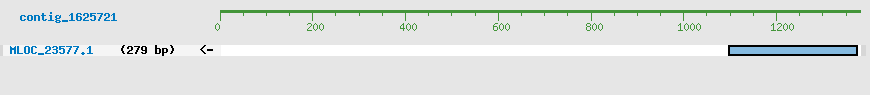
Predicted Genes from contig_1625721
CDS Structure of MLOC_23576
CDS Structure of MLOC_23577
Contig Sequence (1380 bp)
>contig_1625721 1380 assembly3_WGSMorex_rbca.fasta
AGCGGCCCTCGTCGGCGGTGAAGGAGCTCGTCGAGAACAACATCGACGCCGACTCCTCCA
CCATCTCCGTCACCGTCAAGGACGGCGGCCTGAAGCTCATCCAGGTCTCCGGCGGCGGCC
ACGGCATCCGAGTACGCTCGCCCGCCCGCTTCCTAAATAAACCCTAAACCCGGAACCCTT
AGTGCTGCCGCGGGAGCCTACCAGCGCAAAAATCCTTCAGATGCGAGTTGTTAGTCATGA
TTTCTGTTGATTGCGAGCTGCTAATCCTTAGTGCGTACATGATCCGTTGAGTTAAAGACC
AAACACACAGGACCTGGAGGAGTTGATCTACGAGACCAAAAGGTCCAGACCTTGTGTGTT
TGGTCTCCGTTCCCTTGAGTCAATCACCATGATTTTACATCAAGATGATGTTTCTTCTTT
AACAGGGGATGATGAACAACATGGTAACAAGGAGGAACTTTAACCAGCCTGTTATAGACT
AAGCACTGAAAGCATGTTTGCATTTACATTATCTCGGTCATTCTGTTACAGGGCAGTGCA
TTCGTTATTTGTATGATGCTTATTGTTTATTAAAACATCTGTTCCAAGGAATAGATTCTG
ATCTTTTTAGACTATGCTTTTTTTAATGAAAAGTGGAATTACTTGTAATGATAACCTTTA
TAAGGTGGACTTTTATGGACGTTGGAAGTTTTATAATAATGGTTGTTGTACATATGTTAT
CATCATACGAGAGATAATATATTCTTTTGATCCTCACAAACTGTTGTTGCAGATGGAGCT
CATTTTGGCTGAGTTTTCTACCGAGTTGCCACATTTGAGGGCATGCAATTGAAGAATACA
CCACTTTTTTCCAACGATACTAGAGTAATCATATCTAATGAAAGAGAGATGTATTTTATT
TTTGTTGGAGATATTCTGCATCTTAATTTTATGGACAGTAACATTTTAGACAATAATTTG
ATGTCCTAACCATCAAGATCAACTTCGGCATGTATGGTCTCTAGAAGTTACTATTGACCA
TCATTGGTGCATCTTGTAAGGTATAGTCGGTACTATGAGTAACTTACTAGACATGCATTG
ACTGTAGAAGTTGATATACAACTCTTGTGAATTCTAACCATGTTATTCATTTAGGATATA
CACCTAGAAGTGTGTGGGCGTGATTTACAACTCACTAACAGAGCTGCTCAGGCTCGCGAC
CGAGATGAACGTCCTCGGCGATGGTGGTGCCGCGTGTGAGATCCTCCTCCCTAACAAGGC
CGTTGCTGCATCATCTACTCTCTGCCATTGGATCCCCTCCTCTCCGACAAGGCCACCATC
GGATCCCCTCTTCTCCGACAAGGCCATGGTGAGCACCCCTCCGGCCCAGTCCGCCTCCCC



