| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
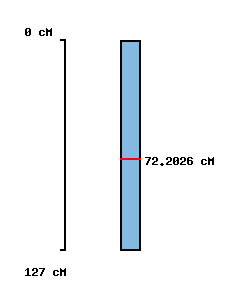 |
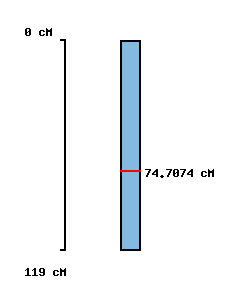 |
No map location |
Predicted Genes from contig_161448
CDS Structure of MLOC_22943
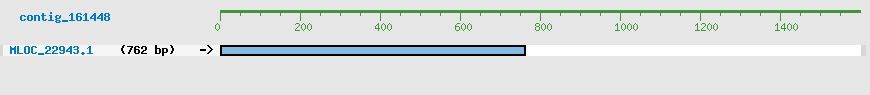
Contig Sequence (1598 bp)
>contig_161448 1598 assembly3_WGSMorex_rbca.fasta
CTCGGCAACGGCGCGCGACGTCCCGCGCGCCGCCGAGGAGGGCGCGGTGAGGCGGCCGGA
GACGATCCAGGAGGGGACGGTGCTCATCCCGGGGATCGGACGGTACGAGCTGGGCAGCCA
CTACAGGCCGGACATCGGCGGGCTGGACCACAGCATCCCGGCCGCCGCCCACGGGCAGTT
CATCCCCGGCGCCGACGACACCTGGGTGCCCAACCCCGGCTTCGAGGTGCCCAACCCATT
CCGCCCCGCAGCCACCACCGAGTCCCCCTGATGGGTCGGTCAAGCCGGCCGGCCAGCACG
CCGTCCGTCCGCGTTAGACCCATCGACCGATGGGTAGCTACTAGCCTGCCATGAACCAAT
CTCCGCTAGCTTTTTTTGGTTGCTACAATAAAATACTAGCTGCTACACGTACGTAGGCAA
GGCATGCGTATGCATGAACCAGCGGCCGCGTGCGTGCGACCGCGCGCGCTGTCTCGACGT
TTTGTGTGTGATTCAAGAGTGTACCCGTATGGGGTATGCGTGGATGCGTAGGAAGGAAGC
ATCCATCGCATATCTCGCTTTGGTTTGTTGTTGCCAATGATGTGTTTCAGTTCACTGAGT
TTCTTTTTAGTTCAATATAATTAAGGTGTTGATGAGGAATTATGCATGATGAAGCTTGCT
ACGTTTGCCAAAAGAAAGAAAATACTTGCATGTATGAAAGGAGATCCATTGTTTGGTACA
GTCATAGTGAGAGTCTGAATTTGATCCTCACTTGGAAGGGACATTATTTTCGGGAGTGAA
TTGCAAAAAACATCTCCATTGAAAGCTAGGTTTGCAGAAACTACACTTCAATGATTTTGT
GAAAAAATCATTAATTACGAAGCTTTTTTTTTTTTGTAACAAAAAAACACTAGTGGTAGC
TTAGGCCGTTTTAAGCATGATTATGACATGTGGGTCCCGCTGAATAGGAAGGCATGGAAA
AAAGTAAAGGCCAATTATTTCAAGGTACAATCTTGTTTTGAAAAACTACCCATGAAAAAA
AGCAATCGGGTCCTTGGCCTTGAAAACAAAAGGCAATCGGAACTTTCGTCTCCCACCGGT
CCCATGGGATTGCTCCGGTGAGCATGTGTCGCTGCCTGACACGTCTAATGGGCGTCTCGG
GAGCGCGAGCACGGACGCCCGCCGCCACTCCCTGCAACTCAGACGGATAGGTCATCCCTG
GCCACCGCGCCACCAAAGCTCGCATAGCTTCGGCTCGGTTGTGAGTCAATGCTGAGGAGG
CCATTGTTTCTTCCTGGAGGTGGCCGACGGGGAGTGGTGGCGTGGGGGTGGAGCTAGAGC
GCGTCGCTCCACCTTTTTTTAAAAGATGACTCCTCTCTTAGTCTTTTTTTACACTTTAGG
CTTTGTTATTTTGACTACTCGTTATGTCGTGTCACTTAACCATGTTCAGCATGACCCATC
TATTATAATCGCGCTCAAAACGGCCTAAGCCACTGACTAGTTGAAAGGACGTGGATGTCG
CCAAGAAGGGGGTGAATAGGCGCTTTAAAATAATTATGGTTTAGGCTTAAACAAANGCGG
AATAAAACTAACGATTAATTTTTCAAGCACAAAACCTA



