| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
| No map location | 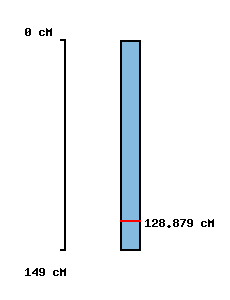 |
No map location |
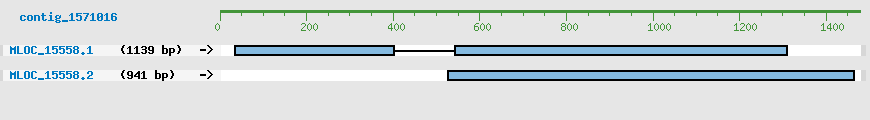
Predicted Genes from contig_1571016
CDS Structure of MLOC_15558
Contig Sequence (1478 bp)
>contig_1571016 1478 assembly3_WGSMorex_rbca.fasta
CGTATCGCATACATTATCCGAAGTTGTGCGAAGGGGCGGGGCGCTCCCCGTGGTGGACCT
GGCGCCGTTCTTCGCCGCGGGAGACGCCGACGACGCGGGCGCGCGCGCCCGTGCCACAGA
CGCCGTGCGCGAGGCCTGCCAGGCCACCGGGTTCTTCCGCGCGGTGAACCACGGCGTGCC
GCCGGAGCTCATGGCGCGCGCTCTCGATCTGTCGGCGGCGTTCTTCGCGCTCCCGGACGA
GGAGAAGGCCAAGGTGCGGCCCGCCGAGGGCGCCTCCCCCGCGCCGCTCCCTGTGGGTTA
CGCGCGGCAGCCCGCGCACTCCGCCGACAAGAACGAGTACCTGCTCCTCTTCAACCCCAA
GCTCGGGCTCAACCACTACCCCGCCCACCCTGCCGGATTCAGGTTACTTCTTTCTTACTC
TGCTTCATTCGTGGATTGTGATTGTAGTTCGTAGCAACCTCTGCTTCGATTCTGATTTCT
GAATTGTGATTCTGAATCCTTCTTATATGAATTGAAAGAAAAAAGAATCTATCATCTTCA
GGGATGCGCTGGAGGAGTGCTACGGCAAGCTCACCGAGCTGGGGCTGCTCATCCAGGACA
TCCTGAGCGAGTGCATGGGCCTGCCGCGAGGCTTCCTCGCCGATTACAACGCCGACCGCG
GCTTCGACTTCCTCACGGCGCTGCGCTACTTCCCGGCGACATCGCCCGACGAGAACAACG
GCATCAGCGCGCACGAGGACGGCAACTGCGTCACCTTCGTCCTGCAGGACGGCGTCGGGG
GCCTGGAGGTGCTCGGGGAAGACGGCCGCTGGGTCCCCGCGGAGCCCGTGGAGGGCAGCA
TCGTCGTCAACGTGGGCGACGTTCTGCAGGTGCTCAGCAACAACAAGTTCAGGAGCGCCA
CGCACAGGGTGGTGAGGAGGCCGGGGGCGCACCGCCACTCCATCGCCTTCTTCCTGAACC
TCCACGGCGACAAGTGGGTCGAGCCGCTGCCGGAGTTCGCGGGCGGCGCCGGCGAGCCGC
CGCGGTACAGAGGGTTCAGGTATAATGAGTACATGCAGCTGCGCATGAGGAACAAGACCC
ACCCGCCGTCCAGGCCCGAGGATGTCGTACACATCACCCATTACCAAATCTAGCTCGCGA
AAGCTTGCTTCCATGATCCATGGCGTACTCTACCTTTTTTCCGTTGTCAAACTTCAGTTC
GTTGTCTGTGGTGTGAACCTAGCCTGTTTTCTGCTGTTGCCTAAGGGCATCTACAATCGG
AGTCCCAGATCGCAGTGGGAGCTCGTATACATGCCGGAGTCAAGCTGGCAATGGTTAATC
CTTATCCGTGTCCCGTGCATAGTTACTCTATCAGGAATGGGGATGTGTCAATTTTAGTGT
TCATGAGAAAGCTGGTGTGTGCGAAATTCCTTGTCGGGTAAGTTCCGACCCGCACGAAAG
GCGTAACGATCTGGGCACTGTCTCGGAGAGAGGCTCGG


