| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
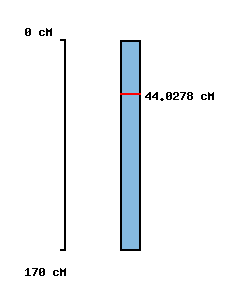 |
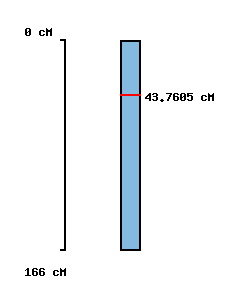 |
No map location |
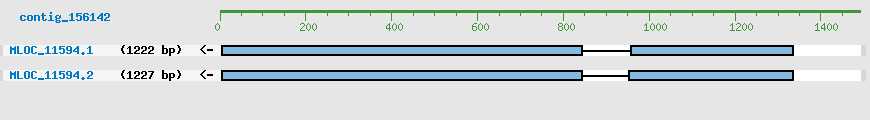
Predicted Genes from contig_156142
CDS Structure of MLOC_11594
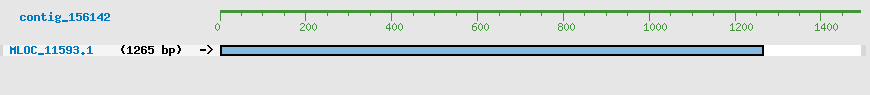
CDS Structure of MLOC_11593
Contig Sequence (1491 bp)
>contig_156142 1491 assembly3_WGSMorex_rbca.fasta
TGTTTGTTGGATAAGAAGGTACAAAAACTAAACGGTGTTCTCATGGCTTTTCTTCTTTAT
GTTTGGTGTTCAAGATACTTATGCCACCATGTAAAATATTTATCTAATAACTTCTCTCTT
GTTGTGCAGCTATATCGGAACCTCTTGATAATGCAGTAGATGAGCTTTGTAATATCTTCT
AACATCTTATATGAACTAGATACTCGCTAATTTAAATTGTTTGATCATACTCAAACGCTT
GAAACACGAAGTATTCTATTGTAAGCTGAAAAATTGTGATGTCACATTCATGTTTCTCGA
AAAACACTCTATACTGCTATCACATTTATTTGTTTATTTTGTTGCATTTGTGTATTATAA
CTCAATGCTCTGTTGATCAGCGTGTTGGTAACTTCGCTCCCTAACACATGCCTCATCTCT
GATGGCGGCGACATGGAACAACACTTCTGTGCGATTAGTACTGTAACTTTGTGAACTGAA
ATTATTATAACCAGTGTTACAAATGTTTGCCGAGGAAAAGTCCTTCCAGCAAAAGAAGAT
CCGGAGGCAGGTTTCCTATCTTGACCAGTATTTCCTTTTGTTCCTAATTGTTGGCTCTTT
CCTATGGAAGCTCTGGAAAAAAATGAAGTGTCCTAAAATTAAGGACTAATGCTTTTGACA
TCTGATTTGAGAAAACTGATTTACAAAATATAACTAGAAGGCAGATTGTAGCATTCATCA
TTGCTATCTTATACAAAGACAGTTGGCGTCTAGAGTGACGCCGTCGACGAGCAAGTCGGC
GTCTGTGAAGCTTCGCTGGAGCCATTCCAGCTCTTCCAGAGTCATAAGGAGAGCCTGAAG
TCAGGGTATGTTCAGTCGAGCACACTCGCTTCTTTCAGAATTTATATATAATCATTAGAA
GTTCAGAACCAGTCATGCCAAAGATATGAAGTGGAACACATTTGCATGCAGCGAAGCTGT
GGAGCCAGGACACCAACGGTGCTAAGCCATTTTCCATCGAGAGGACAATGGCTGCCATCA
GGGACCTCCAGACCAAGCTCAAGATCTCCCTCGCCTCCGTTGATGCCGTGTTTCAAGAGG
GTTATCACCATCCACGACGGCGAGCTAATGCGGTAGCTCGTGGTGCTGATCTGAATGTAT
ATGTTTCTGTTATAGTGTGCTCGATCTAATGATGTATCTTATGTTGCCAAACAACAAAAT
GCTCCTGCCGATGCTTGCATGTGATGACATGGCATTTCAGTTTCTTCCTCGCATGATGTT
CCAATGTAATAAGTCAATCGAGAATTGCATGTACATGTGCTTCTATCAGATAAAAAAATG
AAAGTGAAAAAGGACGTTCATCTACCTGGCACCGCCCATATGCCCCTCAGGCGGCGCGCC
ATGGGCGCGCCCAAACAACTAGTATTCATACTAGTGGCGCACCACTCAGCACTTTAGTGG
CGCACTTCAAAAAGCACACTAGTGGCACATCGTCAACTAGTGCGCCATTAG




