| Morex x Barke POPSEQ 2013 | Oregon Wolfe POPSEQ 2013 | Morex Genome Release 2012 |
|---|
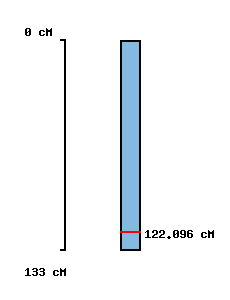 |
No map location | 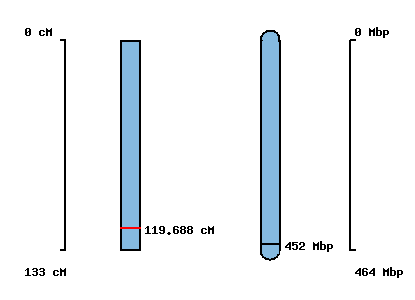 |
Predicted Genes from contig_148569
CDS Structure of MLOC_9033
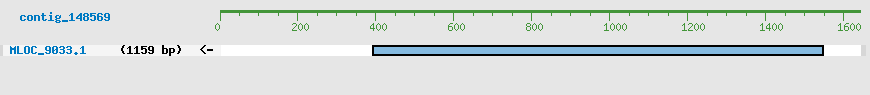
Contig Sequence (1642 bp)
>contig_148569 1642 assembly3_WGSMorex_rbca.fasta
GAGGTCGGCCGTGGCGGCGAGTGGTGTGGCGGCGGCCTGGGCACATGCCAGGATGGCCAA
GGGTCGACGGGAGGAGGCGATGCGTACGGATATAATTTTTGCAGCGAATCGTTTTTTCCT
TTTGCGTTGTAGATAAATGATGGAGCGCGGGTTGAATAACAAAAATTACAGGGACTTTTT
ATAGAAATATCGTGGTGGGTTTTTCCGACGAAAGCAATAGCCGCTTTATTGTTAGGTATA
GATTTTGCAGACTGCTAGGTCTGCATGTCGCTGACTGTCCAAACCGTACAATATAAACGC
TTAAAGATCTGAACAGATTGATGCAGTCCTGTGAAGCTAGCACATATCTTGCTTTGAGGC
GAGTACTTGCTCACATGCTTTTGTTTCAAATAAAAACGAGAGGAAAGTGTTAGACATGCT
AAGCTCAGGTCACTAGGCTAGTTTTCGGCTTAAAATAAATAGTGCGCACTAGGCTGCGGA
TGCGACACTAGTAGATGGAGAAGAAAATGCTGCGCACTATCTTCTAACATTCGTGGACCT
GGAATGACTATGCATAGATCATCACTTTTTGTGTGGTCCGTTCTAGTGACAGCATTCCTA
CTTTTATTATCACTTCCGGTACTGGCGGGGGCAATTACAATGTTATTAACCGATCGAAAC
TTTAATACAACCTTTTTTGATCTTGCAGGAGGGGGAGACCCCATATTATACCAGCATCTC
TTTTGGTTCTTCGGTCATCCAGAGGTGTGTATTTTCATTCTGCAGTTACGGCAGCATCAA
AACTACTCATCTCTCGCACGTTCATCTTTCTCGCGAGTTCTCTCTCTCGCTGATCCGGCG
ACAACAAAAAGTGTGGCCATGCGAGTTGTGACTGTTGCAAAACTAAAGCCTGAGCCCAAC
AAATTCACTAGCCAATATGCCCCATAAGTTTGTCGCTCATCTAAGAACTGCGCGGCAATG
GTTTTTTCTGATAATCCCAACAAATTGCTAGATGCCAAATCTAACAATGCTTCTCCAGAG
AATTCAATTCTCATAATTTACATCATGGTAGTACAAATCAAACCACACAACCTCACACTT
CACTGTGAGCAGCTCCAGAACACCTTATTTGCCGGGGACGAAGTTGGTGGCAAATGCCCA
GGCGTTGTTGTTGACAGGGTCGGTGATGTGGTCACCAAGGTTCTCGAGTGGGCCCTTGCC
GGTGACTATGGCCTGGACAAAGAAGCCAAACATTGAGAACATGGCAAGGCGGCCGTTCTT
GATCTCCTTCACCTTAAGCTCGGCGAATGCCTCGGGGTCATCAGCCAGACCGAGTGGGTC
AAAGCTTCCGCCCGGGTAGAGAGGGTCAACGATCTCGCCGAGTGGGCCGCCGGCAACACG
GTACCCCTCAACAGCGCCCATGAGCACAACCTGGCAAGCCCAAATGGCAAGGATGCTTTG
AGCATGGACGAGGCTTGGGTTGCCGAGGTAGTCGAGGCCACCCTCGCTGAAGATTTGCGA
GCCGGCCTTGAACCAGACAGCCTCGCCGAACTTCACGCCATTGCGGCGGGCAAGCAGCAA
CAACTCAGGGAACACGCAACCCAGTGCACCAAGCATAGCCCAGCGGCAGTGGATCACCTC
CAACTCGCGGTTCTTCGAGAAG



