Matching HORVU genes
BarleyRTD transcripts were compared to the location of the IBSC 2017 Predicted Transcripts. Overlapping status is classified as Containing (HORVU is smaller than the BART), Contained (BART is smaller than the HORVU), Partial-Overlap (Overlap exists), and Undefined (the BART transcript has no overlapping HORVU).
| HORVU ID | Percentage overlap | BART Gene Direction | HORVU Gene Direction | Overlap Length | Gene Length Difference | Overlapping Status |
|---|
| HORVU3Hr1G061690 | 83.6 | + | + | 3965 | -781 | Containing |
Transcripts from Gene BART1_0-u21412
Exon Structure of BART1_0-u21412
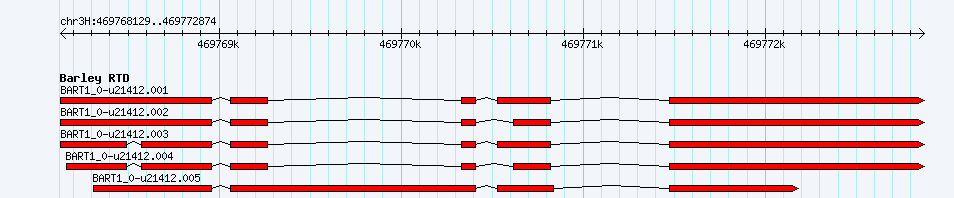
grey: non-coding,
green: Barley Morex IBSC 2017 Transcript CDS,
red: Barley RTD exons
Homology of Longest Transcript to Model Species (BLASTX to E-value < 1e-30)
| Database | Hit | Frame | E-value | Score | % Identity | Description |
|---|
| Rice PP7 |
LOC_Os01g48190.1 |
+2 |
4e-60 |
206 |
124/163 (76%) |
protein|drought induced 19 protein, putative, expressed |
| TAIR PP10 | None | - | - | - | - | - |
| BRACH PP3 |
Bradi2g46240.1.p |
+2 |
9e-71 |
236 |
144/166 (87%) |
pacid=32778880 transcript=Bradi2g46240.1 locus=Bradi2g46240 ID=Bradi2g46240.1.v3.1 annot-version=v3.1 |
GO Annotation
Generated by PANNZER
Barley PseudoMolecules GBrowse
Click here to see more tracks within GBrowse -->
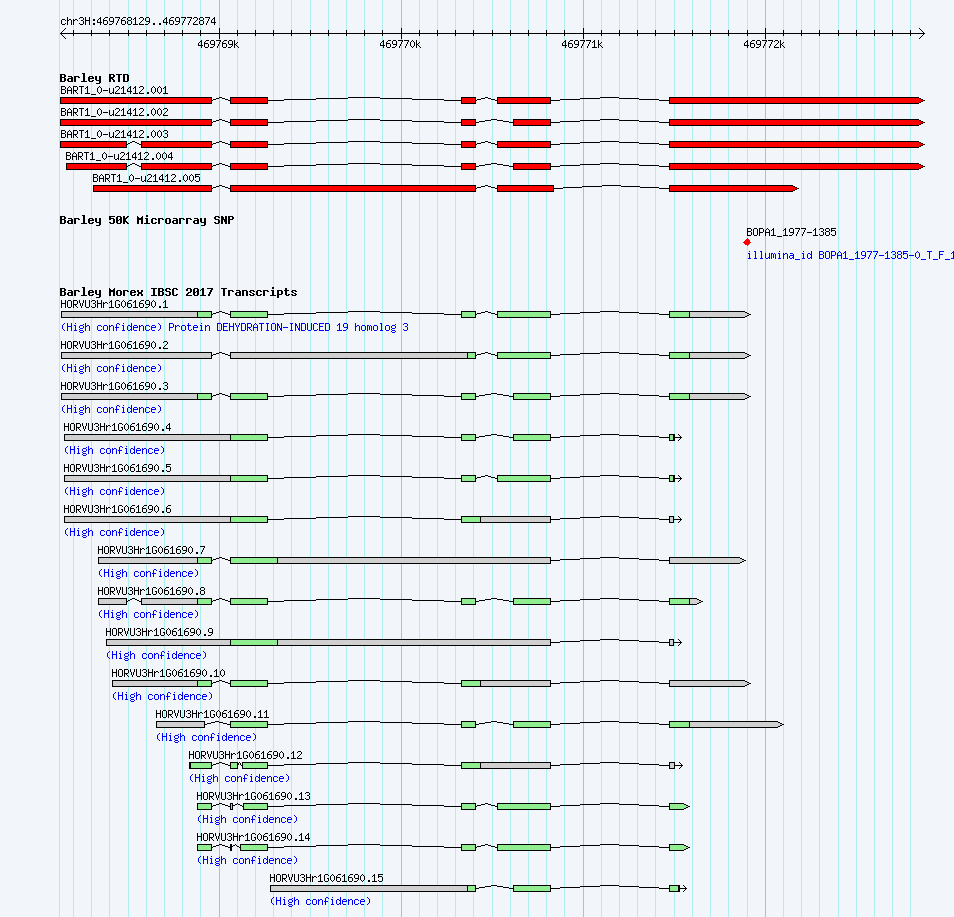
grey: non-coding,
green: Barley Morex IBSC 2017 Transcript CDS,
red: Barley RTD exons
